Deriving life history traits from an MPM
Patrick Barks
Judy Che-Castaldo
Rob Salguero-Gomez
Others
2025-09-15
Source:vignettes/a03_LifeHistoryTraits.Rmd
a03_LifeHistoryTraits.RmdIntroduction
The aim of this vignette is to provide further details on the
functions in Rage for estimating life history traits and on
the traits themselves. Life history describes the sequence and pattern
of the key events in an organism’s life cycle, pertaining to the
schedule of survival, development, and reproduction. By aggregating
individual-level demographic rates into an MPM, we can calculate a set
of life history traits to describe the expected patterns for individuals
within the population. These calculations follow methods from Caswell
(2001) and Morris & Doak (2003).
Loading MPMs and basic anatomy
We will start by loading the Rage package and an example
MPM included in the package called mpm1, which we’ll be
using throughout this vignette.
library(Rage) # load Rage
data(mpm1) # load data object 'mpm1'
mpm1 # display the contents
#> $matU
#> seed small medium large dormant
#> seed 0.10 0.00 0.00 0.00 0.00
#> small 0.05 0.12 0.10 0.00 0.00
#> medium 0.00 0.35 0.12 0.23 0.12
#> large 0.00 0.03 0.28 0.52 0.10
#> dormant 0.00 0.00 0.16 0.11 0.17
#>
#> $matF
#> seed small medium large dormant
#> seed 0 0 17.9 45.6 0
#> small 0 0 0.0 0.0 0
#> medium 0 0 0.0 0.0 0
#> large 0 0 0.0 0.0 0
#> dormant 0 0 0.0 0.0 0Although we demonstrate these trait calculations using a single MPM, each function can be applied to a set of MPMs for comparative analysis of life histories across species and populations.
Survival and lifespan traits
Two life history traits pertaining to survival that can be estimated
using functions in Rage are mean life expectancy and
longevity. Assuming that there is no trade-off between survival and
reproduction, these functions do not require information about
reproduction. Instead, they only require a U matrix
(supplied to the functions’ matU argument).
The function life_expect_mean() estimates mean life
expectancy, or the mean time to death, and the variance in time to death
can be obtained using life_expect_var().
# mean life expectancy from "seed" stage
life_expect_mean(matU = mpm1$matU, start = 1)
#> [1] 1.250506
# variance in life expectancy from "seed" stage
life_expect_var(matU = mpm1$matU, start = 1)
#> [1] 0.7816213Life expectancy is dependent on a starting stage, which is specified
using the start argument. That is, the expected time to
death will be different for an individual that is currently in the first
stage (i.e., start = 1) than if it is in a later stage
along its life cycle. This is because the latter assumes the individual
has survived to that stage, and it has already lived a portion of its
lifespan. In the example MPM, life expectancy from the first stage
(“seed”) is shorter than that from the second stage (“small”) due to the
relatively low probability of survival in the seed stage, reducing the
expected time to death.
# mean life expectancy from "small" stage
life_expect_mean(matU = mpm1$matU, start = 2)
#> [1] 2.509116In certain situations, it is useful to compute life expectancy while
taking into account that there can be multiple initial stage classes
rather than a single stage from which every individual commences their
life cycle. This need occurs in scenarios where individuals can enter a
life stage, such as reaching reproductive maturity, from more than one
possible stage. To accommodate these instances, the
life_expect_mean function supports multiple initial stage
classes. In these scenarios, results should be averaged, with weights
based on the distribution of individuals among the potential starting
stages. This is achieved by setting the start parameter to
NULL, which signifies an unspecified starting stage, and
employing the mixdist parameter.
The mixdist parameter is expected to be a vector
reflecting the proportion of individuals initiating reproduction at each
stage within the Matrix Population Model (MPM). By inputting this
distribution, the function computes the life expectancy by weighting the
contribution from for each initial stage accordingly.
life_expect_mean(matU = mpm1$matU, start = NULL, mixdist = c(0, 0.4, 0.6, 0, 0))
#> [1] 2.865667The function longevity() estimates the time to which the
survivorship of a cohort falls below a user-defined critical threshold,
specified as a value (between 0 and 1) supplied to the
lx_crit argument. The specifications regarding the
start argument for life_expect() also applies
to this function. Using the example MPM, the post-germination years
until survivorship falls to below 5% would be calculated as:
longevity(matU = mpm1$matU, start = 2, lx_crit = 0.05)
#> [1] 7Similarly to the mean life expectancy function, here one too can examine how longevity differs depending on the starting stage. With the example MPM, longevity increases from 2 years to 7 years after an individual germinates. Longevity is highest for the “medium” and “large” stages, but is the same for individuals starting in the “small” stage as those starting in the “dormant” stage.
longval <- NULL
startvec <- seq_len(nrow(mpm1$matU)) # vector of starting stages
for (i in startvec) {
longval[i] <- longevity(matU = mpm1$matU, start = startvec[i], lx_crit = 0.05)
}
plot(longval,
type = "l", xlab = "Starting stage",
ylab = "Longevity to 5% survivorship"
)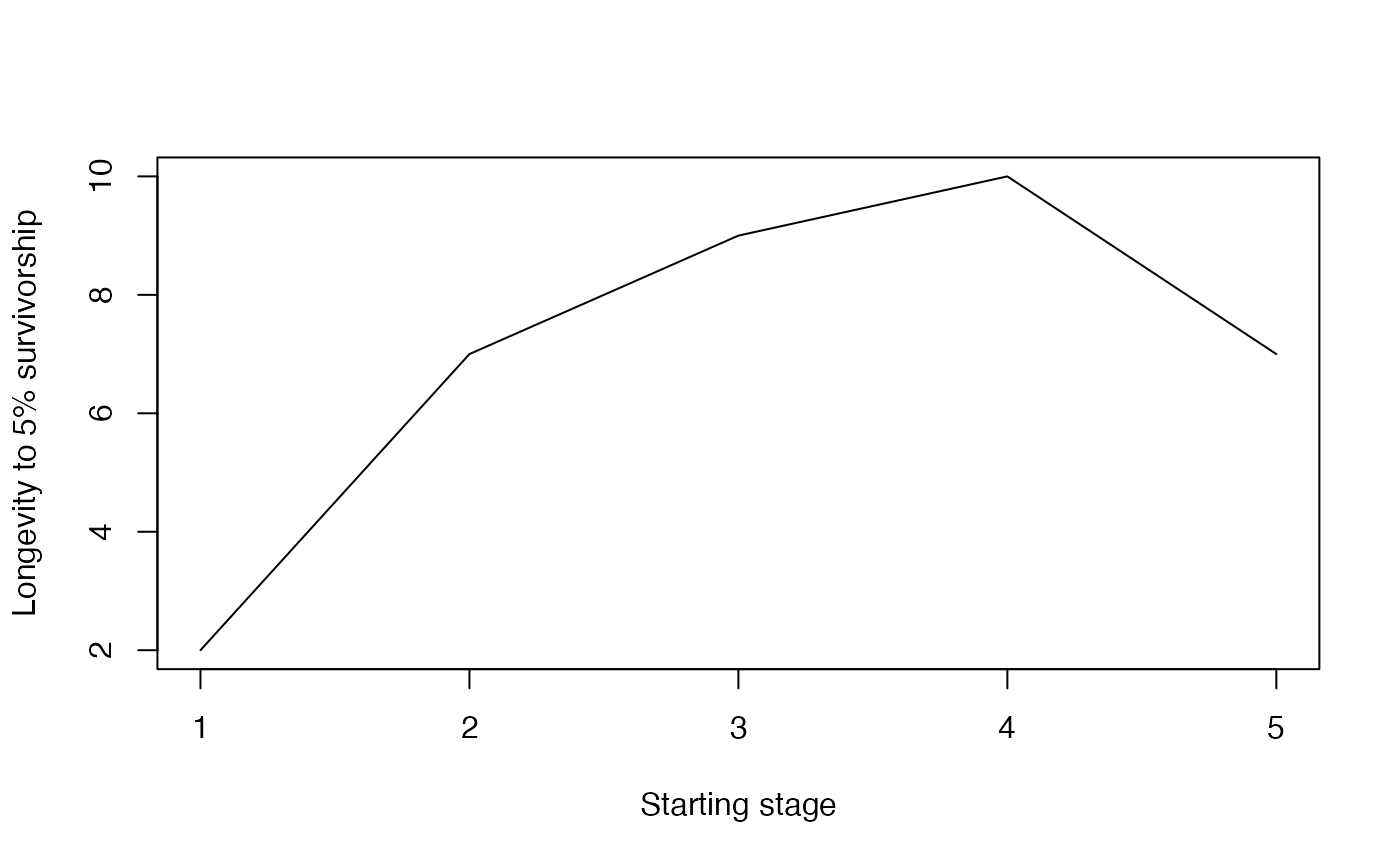
longval
#> [1] 2 7 9 10 7Reproduction and maturation traits
In addition how long individuals can be expected to live, species and populations also differ in how quickly and often they reproduce, and how many offspring are produced and recruited into the population. Several related traits measure these various aspects of reproduction, and they require information contained in the F submatrix as well as the U submatrix.
The net reproductive rate, R0 is the cumulative number of offspring that a newborn individual will produce over its lifetime.
net_repro_rate(matU = mpm1$matU, matR = mpm1$matF)
#> [1] 1.852091This life history trait is also called the the per-generation growth rate, because if an individual produces more than one offspring, it more than replaces itself and so the population will grow. Note that this growth rate is different than the population growth rate $because the units of R0 are in generations rather than absolute time (i.e., the time step of the transition matrix, often 1 year). The generation time (T) can thus be defined as the amount of time it takes for a population to grow by a factor of R0, which can be obtained by:
gen_time(matU = mpm1$matU, matR = mpm1$matF)
#> [1] 5.394253Generations may be bounded using different criteria, leading to
estimates of T that are identical only for organisms that
reproduce once at a uniform age. Alternative methods for calculating the
generation time can be selected using the method argument.
Currently supported are (i) the average age difference between
parents and offspring (method = "age_diff") and
(ii) the expected age at which members of a cohort reproduce
(method = "cohort").
The age at reproductive maturity is the average amount of time an
individual will take to enter a reproductive stage for the first time in
the population. Similar to life expectancy, mean time to reproduction
will depend on the starting stage, again specified using the
start argument. In the case of the example MPM where the
first stage is “seed”, one may be more interested in estimating the time
to reproduction for individuals that have already germinated (i.e.,
start = 2).
mature_age(matU = mpm1$matU, matR = mpm1$matF, start = 2)
#> small
#> 2.136364The probability of reaching reproductive maturity before death, for individuals that have already become established (e.g. germinated in the case of plants in this particular example), is:
mature_prob(matU = mpm1$matU, matR = mpm1$matF, start = 2)
#> [1] 0.4318182As discussed above, there may be multiple stages in which an
individual can first reproduce. One can also use an MPM to estimate the
proportion of individuals that would be expected to first reproduce in
each of the reproductive stage classes. For this, we need to specify
which stages are reproductive in the repro_stages argument.
This can be determined from the F matrix based on the
columns that have non-zero values, or a non-zero probability of
producing offspring.
mpm1$matF # We see that the "medium" and "large" stages are reproductive
#> seed small medium large dormant
#> seed 0 0 17.9 45.6 0
#> small 0 0 0.0 0.0 0
#> medium 0 0 0.0 0.0 0
#> large 0 0 0.0 0.0 0
#> dormant 0 0 0.0 0.0 0
maturedist <- mature_distrib(
matU = mpm1$matU, start = 1L,
repro_stages = c(FALSE, FALSE, TRUE, TRUE, FALSE)
)
maturedist
#> seed small medium large dormant
#> 0.00000000 0.00000000 0.92105263 0.07894737 0.00000000One can then use this distribution to update the estimate of mean life expectancy from reproductive maturity, using the data-based distribution rather than the hypothetical one (40% in “small” stage and 60% in “medium” stage) used above:
# mean life expectancy from maturity
life_expect_mean(matU = mpm1$matU, start = NULL, mixdist = c(maturedist))
#> [1] 3.179005
# mean life expectancy from "small" stage
life_expect_mean(matU = mpm1$matU, start = 2)
#> [1] 2.509116In doing so, mean life expectancy from maturity is 0.67 years longer than that after germination (from “small” stage). In this case, this difference is due to the higher average survival once an individual reaches reproductive maturity.
Life table component traits
Other life history traits are calculated from a life table rather
than an MPM, in which case one can first use the mpm_to_
group of functions to derive the necessary life table components:
age-specific survivorship (lx), survival probability
(px), mortality hazard (hx), and reproduction
(mx). As above, start stage can too be specified as a specific
stage (by number or by name), as well as a vector of proportions in each
stage. One can calculate the age-specific rates for post-germination
individuals using the example MPM:
lx <- mpm_to_lx(matU = mpm1$matU, start = "small")
px <- mpm_to_px(matU = mpm1$matU, start = "small")
hx <- mpm_to_hx(matU = mpm1$matU, start = "small")
mx <- mpm_to_mx(matU = mpm1$matU, matR = mpm1$matF, start = "small")There are multiple applications for these life table estimates. For example, one can visualize the difference in survival curves for individuals starting from the seed stage compared to survival post-germination.
lx_seed <- mpm_to_lx(matU = mpm1$matU, start = "seed")
plot(lx,
xlab = "Survival time (years)", ylab = "Survivorship",
type = "s", col = "black"
)
lines(lx_seed, type = "s", col = "orange")
legend("topright",
inset = c(0.05, 0.05), c("From seed stage", "From small stage"),
lty = c(1, 1), col = c("orange", "black")
)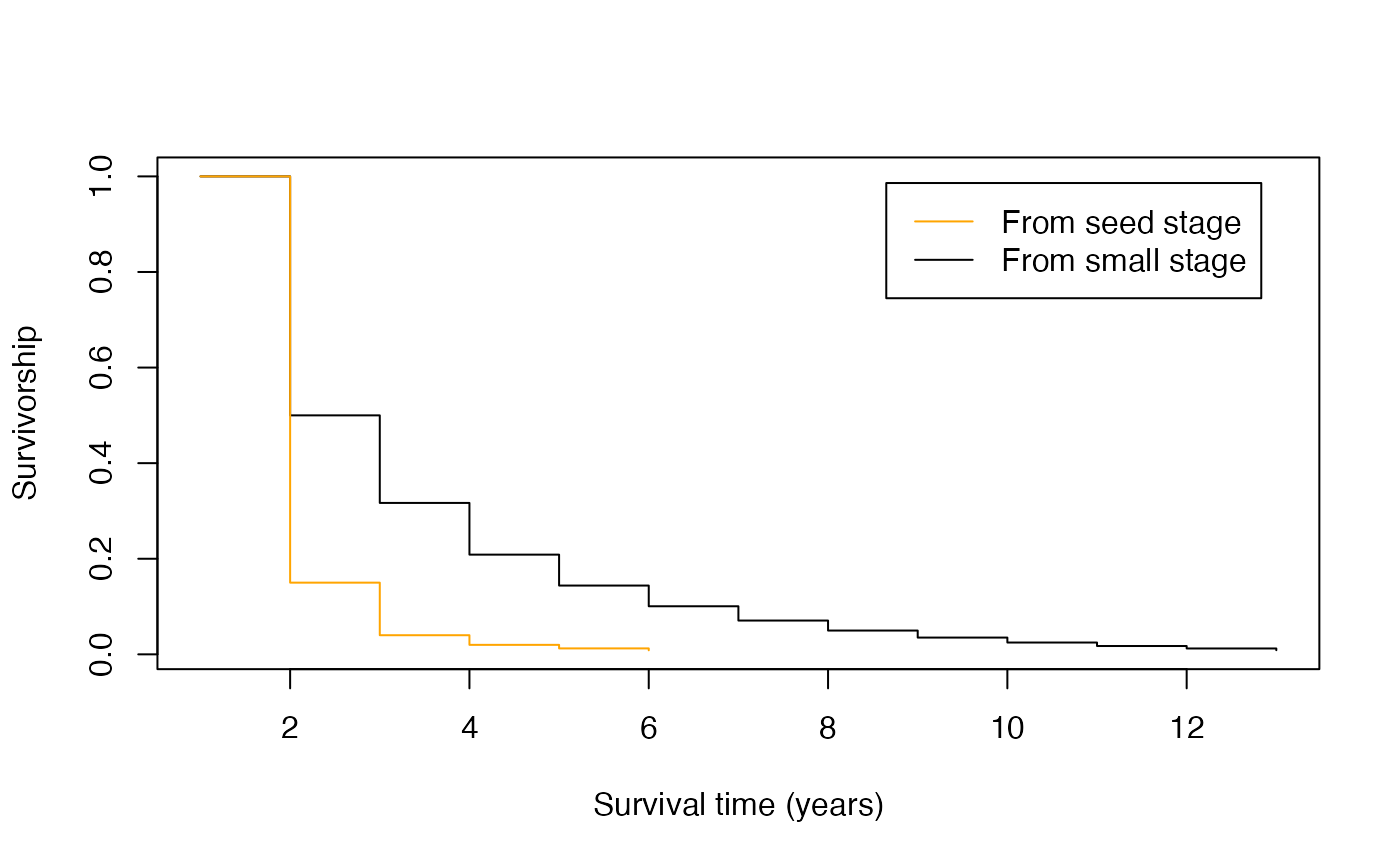
In this case, there is a large drop in survival (and thus high mortality) in the first year for seeds. Specifically, more than 80% of seeds are not expected to survive their first year, compared to germinated individuals that have a 50% chance of surviving their first year.
Finally, one can use these life table components to calculate additional life history traits related to the types of survival and reproduction trajectories.
Demetrius’ entropy is a measure of how reproductive episodes are spread across the lifespan (Demetrius & Gundlach 2014). A value of 0 indicates only one reproduction event in the entire life time (also known as semelparity), and larger values indicate a more even distribution of reproductive events over the life cycle.
Keyfitz’ entropy describes the shape of the age-specific survivorship curve (Demetrius 1978). Values greater than 1 correspond to survival curves type I (high survival in early part of life and decreasing over time), a value of 1 to survival curve type II (constant survival), and values less than 1 to survival curve type III (low survival early and increasing over time) (Salguero-Gómez et al. 2016).
These can be calculated from the age-trajectories, or from the matrices directly (in this case, the age-trajectories are calculated internally).
Recently, de Vries et al. (2023) highlighted a problem in the
calculations of Keyfitz’ entropy for discrete age trajectories.
Following this revelation it is recommended to use the function
entropy_k_age to calculate entropy for age-based (Leslie)
matrices and entropy_k_stage for stage-based Lefkovitch
matrices.
entropy_d(lx, mx) # Demetrius' entropy
#> [1] 0.7735551
entropy_d(lx = mpm1$matU, mx = mpm1$matF, start = "small") # Demetrius' entropy
#> [1] 0.7735551
life_elas(lx) # Keyfitz' entropy
#> [1] 0.9077186
life_elas(lx = mpm1$matU, start = "small") # Keyfitz' entropy
#> [1] 0.9077186
entropy_k_stage(mpm1$matU)
#> [1] 1.131567
data(leslie_mpm1) # load leslie matrix
entropy_k_age(leslie_mpm1$matU)
#> [1] 0.626804It is important to note that both of these entropy measures may produce unexpected results if only partial survivorship and fecundity age-based trajectories are used in the calculations. Indeed, both metrics of demographic entropy are sensitive to the length of the those trajectory vectors. For example, the Demetrius’ entropy calculated above has a negative value due to being calculated from age-specific survivorships and fecundities starting from the “small” stage, and therefore it does not include the full life cycle.
A more robust way to describe these trajectories is using the area under the curve method.
Shape of mortality and fecundity
A life course can be described in terms of its pace (timing of
events) and its shape (the distribution of events, either for an
individual or for a population). There are several candidate metrics for
the pace and shape of mortality, and of fertility. A good measure of the
pace of mortality is life expectancy (the average age of death) and a
good measure of the shape of mortality are the life-table entropy
measures mentioned above, which describe both the survivorship curve and
the mortality trajectory (see Baudisch 2011). Measures describing the
pace and shape of fertility have also been developed (Baudisch &
Stott 2019). In addition to the entropy measures described above, the
shape_ functions are also useful metrics.
# shape of survival/mortality trajectory
shape_surv(lx)
#> [1] -0.04681254
# shape of fecundity trajectory
shape_rep(mx)
#> [1] -0.02999578References
Baudisch, A. 2011. The pace and shape of ageing. Methods in Ecology and Evolution, 2(4), 375– 382. doi:10.1111/j.2041-210X.2010.00087.x
Baudisch, A, Stott, I. 2019. A pace and shape perspective on fertility. Methods Ecol Evol. 10: 1941– 1951. doi:10.1111/2041-210X.13289
Caswell, H. 2001. Matrix Population Models: Construction, Analysis, and Interpretation. 2nd edition. Sinauer Associates, Sunderland, MA. ISBN-10: 0878930965
Demetrius, L. 1978. Adaptive value, entropy and survivorship curves. Nature 275: 213-214.
Demetrius, L., & Gundlach, V. M. 2014. Directionality theory and the entropic principle of natural selection. Entropy 16: 5428-5522.
de Vries, C., Bernard, C., & Salguero-Gómez, R. 2023. Discretising Keyfitz’ entropy for studies of actuarial senescence and comparative demography. Methods in Ecology and Evolution, 14, 1312–1319. doi:10.1111/2041-210X.14083
Morris, W. F. & Doak, D. F. 2003. Quantitative Conservation Biology: Theory and Practice of Population Viability Analysis. Sinauer Associates, Sunderland, MA. ISBN-10: 0878935460
Salguero-Gómez, R., Jones, O. R., Jongejans, E., Blomberg, S. P., Hodgson, D. J., Mbeau-Ache, C., Zuidema, P. A., de Kroon, H., & Buckley, Y. M. 2016. Fast–Slow continuum and reproductive strategies structure plant life-history variation worldwide. Proceedings of the National Academy of Sciences 113 (1): 230–35