2 User Guide
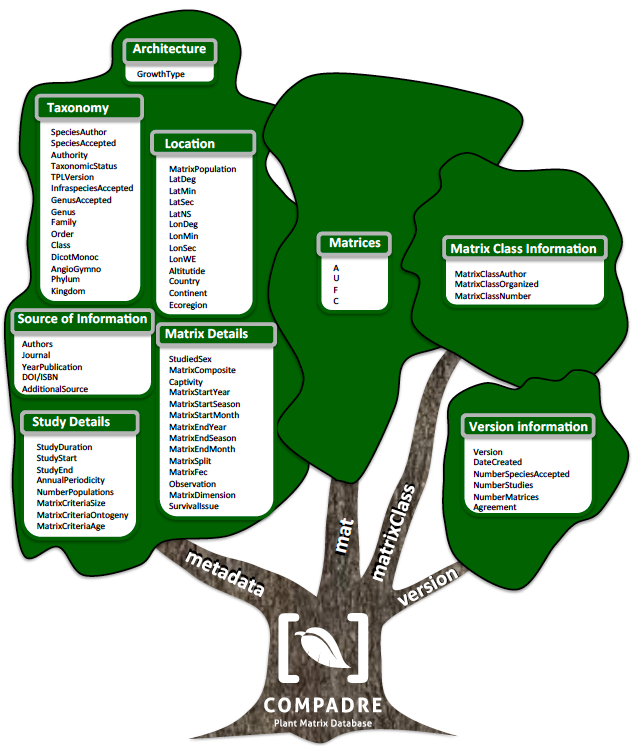
Figure 1: Variables archived in COMPADRE, organized according to the general category to which they are associated. The R data object compadre is a list containing further list and data.frame objects (named metadata, mat, matrixClass and version), here represented by each branch of the tree. The canopy contains the variables in each data.frame/list, organized by general categories.
2.1 Variables in metadata
The object metadata is a data.frame detailing the ancillary metadata that give context to each matrix population model (MPM). These are detailed below.
2.1.1 MatrixID
Definition Unique identification number given to each MPM and its associated metadata
Possible values:
MatrixID- e.g. 238263
2.1.3 SpeciesAccepted (Population form)
Definition: Currently accepted latin name. This information is obtained from GBIF
Possible values:
- Genus species - e.g. “
Taraxacum officinale”, “Icaricia icarioides fenderi”, “Iris pallida subsp. cengialti”, etc.
Note on nomenclature in COMPADRE and COMADRE: Species in COMPADRE are displayed following guidelines in the International Code of Nomenclature for algae, fungi, and plants (ICN) (i.e. abbreviated infraspecific type is displayed between the specific and subspecific epithets), and species in COMADRE are displayed following guidelines in the International Code of Zoological Nomenclature (ICZN) (i.e. abbreviated infraspecific type is not displayed between the specific and subspecific epithets).
Examples:
- “
Anthyllis vulneraria subsp. alpicola” and “Hosta undulata var. undulata” in COMPADRE - “
Panthera tigris sumatrae” and “Somateria mollissima borealis” in COMADRE
2.1.4 CommonName (Population form)
Definition: Common name as used in the publication
Possible values:
- Common_name - e.g.
American black bear
2.1.5 Kingdom (Species form)
Definition: Kingdom to which species belongs
Possible values:
- kingdom - e.g. Plantae, Fungi, Rhodophyta, Chromista (COMPADRE includes fungi and algae as well as plants, and COMADRE includes viruses and bacteria as well as animals)
2.1.6 Phylum (Species form)
Definition: Phylum to which species belongs
Possible values:
- phylum - e.g.
Magnoliophyta
2.1.7 Class (Species form)
Definition: Class to which species belongs
Possible values:
- class - e.g.
Magnoliopsida
2.1.8 Order (Species form)
Definition: Order to which species belongs
Possible values:
- order - e.g.
Caryophyllales
2.1.9 Family (Species form)
Definition: Family to which species belongs
Possible values:
- family - e.g.
Polygonaceae
2.1.10 Genus (Species form)
Definition: Taxonomic genus name of study SpeciesAccepted
Possible values:
- genus - e.g.
Chorizanthe
2.1.11 Species (Species form)
Definition: Taxonomic species name of study SpeciesAccepted
Possible values:
- species - e.g. sapiens
2.1.12 Infraspecies (Species form)
Definition: Taxonomic infraspecies name of study SpeciesAccepted
Possible values:
infraspecies- e.g. fenderi
2.1.13 InfraspeciesType (Species form)
Definition: Infraspecific type of study SpeciesAccepted
Possible values:
infraspecies type- e.g. subspecies, variety, color form
2.1.14 OrganismType (Species form)
Definition: General organism type, based mainly on architectural organization. The species was assigned to one of these possible values using the description of plant growth type provided by the author for COMPADRE; animal taxonomic class, virus, or bacteria for COMADRE; and other external sources (e.g. other publications)
Possible values for COMPADRE:
Algae- A brown, green or red alga. Note that green algae are in kingdom Plantae, but should still be recorded as OrganismType = AlgaeFungi- A fungus species, including yeasts and molds as well as multicellular fungi.Annual- Typically plant ecologists also refer to pseudoannual species as being “biennials”. The main difference between the annuals and biennials is that annual species complete their lifecycle (are born, grow, reproduce and die) within a single year, whereas biennials have the potential to stretch that time-window up to two years. Both annuals and biennials are often associated with periodic (or seasonal) matrices. Biennial (and pseudoannual) species are categorized as “Annual” in COMPADREBryophyte- a bryophyteEpiphyte- an epiphyteFern- a fernHerbaceous perennial- a herbaceous perennialLiana- a lianaPalm- a palmShrub- a shrubSucculent- a succulentTree- a tree
Possible values for COMADRE:
- taxonomic class - e.g. “
Mammalia”, “Aves”, “Gastropoda”
2.1.15 DicotMonoc (Species form)
Definition: Whether species is a dicot or monocot
Possible values:
Eudicot- species is a dicotMonocot- species is a monocotNA- Species is neither a dicot nor a monocot (not an angiosperm) or species in COMADRE
2.1.16 AngioGymno (Species form)
Definition: Whether species is an angiosperm or a gymnosperm
Possible values:
Angiosperm- Species is an angiospermGymnosperm- Species is a gymnospermNA- Neither angiosperm nor gymnosperm or species in COMADRE
2.1.18 Journal (Publication form)
Definition: The name of the journal from which data were collected
Possible values:
- abbreviated journal name - Where the data come from a scientific journal article, the abbreviated journal name is given. We use the standard abbreviation of the journal using the BIOSIS Format
NA- Data sourced from other material
2.1.19 SourceType (Publication form)
Definition: The type of source from which the data were collected
Possible values:
Journal Article- Matrices are from a journal articleBook- Matrices are from a bookBook Chapter- Matrices are from a book chapterThesis- Matrices are from a doctoral dissertation or masters thesisReport- Matrices are from a reportConference- Matrices are from a conference talk or posterOther- Matrices are from another type of source, e.g., website, preprint
2.1.20 OtherType (Publication form)
Definition: Type of source from which the data were collected when SourceType is ‘Other’
Possible values:
Internet- Matrices are from an internet source, e.g. websiteUnpublished- Matrices are unpublished and provided by the data collectorPreprint- Matrices are from an article preprint, e.g. bioRxiv
2.1.21 YearPublication (Publication form)
Definition: Year of publication
Possible values:
- yyyy - e.g.
2012
2.1.22 DOI_ISBN (Publication form)
Definition: Digital Object Identifier or International Standard Book Number codes that identify the publication.
Possible values:
- DOI or ISBN - e.g. “
10.1111/1365-2745.12334”
2.1.23 AdditionalSource (Publication form)
Definition: Additional source(s) used to reconstruct the matrix, or to obtain additional information on ancillary data
Possible values:
- Lastname Journal YearPublication - e.g.
Godinez-Alvarez Bot Rev 2003
2.1.24 StudyDuration (This is calculated using the fields Study End and Study Start; Publication form)
Definition: The number of years of observation. This is calculated as (StudyEnd year - StudyStart year) + 1 (see below). The calculation thus overlooks any missing years in the middle of the study period
Possible values:
- integer - e.g.
14 Simulation- The matrices in the study are from simulated data only and not from real-world or experimental data
2.1.25 StudyStart (Publication form)
Definition: First year of study
Possible values:
- yyyy - e.g.
2012 Simulation- The matrices in the study are from simulated data only and not from real-world or experimental data
2.1.26 StudyEnd (Publication form)
Definition: Final year of study
Possible values:
- yyyy - e.g.
2014 Simulation- The matrices in the study are from simulated data only and not from real-world or experimental data
2.1.27 ProjectionInterval (Formerly AnnualPeriodicity; Matrix form)
Definition: Indicates the time step (periodicity) for which the seasonal or annual transition matrix was constructed, e.g., ‘1’ indicates that the MPM iteration period is 1 year; 0.5 indicates that the MPM iterates once every 0.5 years or 6 months; 2 indicates that the MPM iteration occurs every 2 years
Possible values:
- numeric value - e.g.
1,2,0.2etc.
2.1.28 MatrixCriteriaSize (Population form)
Definition: Indicates whether the matrix population model contains stages based on size
Possible values:
Yes- The matrix population model contains stages based on size (e.g. DBH, stem length, stem height, stem number, etc.)No- The matrix population model is not based on size
2.1.29 MatrixCriteriaOntogeny (Population form)
Definition: Indicates whether the matrix population model contains stages based on development
Possible values:
Yes- The matrix population model contains at least one class that is based on development/ontogeny (e.g. seedbank, reproductive, vegetative, dormant, adult, etc.)No- Matrix population model is not based on ontogeny
2.1.30 MatrixCriteriaAge (Population form)
Definition: Indicates whether the matrix population model contains stages based on age
Possible values:
Yes- The matrix population model contains at least one class that is based on age (e.g. 1, 2, 3, etc.)No- The matrix population model is not based on age
2.1.31 MatrixPopulation (Formerly PopulationName; Population form)
Definition: Name of population where matrix was recorded, usually as given by the publication author. In some cases, where the author provides no name, we give the closest geographic location instead. If there are multiple populations in the study and their names are not pertinent/available, sequential names in alphabetical order are assigned for each population in the study (e.g. “A”, “B”, “C”, etc.)
Possible values:
- any text string - e.g. “
Brazeau Creek, Florida”.
2.1.32 NumberPopulations (This has to be calculated, it is not a field, at least not one that can be observed on our end)
Definition: Number of populations of the study species (SpeciesAccepted) within a publication. This variable refers to the number of populations as defined by the author. Within-site replication of permanent plots are not considered to be different populations (see below)
Possible values:
- integer - e.g.
3(for three populations)
2.1.33 Lat (From the coordinates field. If it was recorded in DMS then it is transformed to decimal degrees; Population form)
Definition: Degree value of latitude in decimal degree format. Negative values indicate that the location is in the southern hemisphere
Possible values:
- -90-90 - e.g.
64.87,-43.7
2.1.34 Lon (From the coordinates field. If it was recorded in DMS then it is transformed to decimal degrees; Population form)
Definition: Degree value of longitude in decimal degree format. Negative values indicate that the location is to the west of the Greenwich meridian (0 degrees)
Possible values:
- -180-180 - e.g.
137.78,-78.32
2.1.35 Altitude (Population form)
Definition: Altitude of studied population defined as height above or below sea level in meters of the population. Negative numbers indicate populations below sea level
Possible values:
- numeric value - e.g.
653
2.1.36 Country (Population form)
Definition: Country/ies where study took place. We use ISO 3 three letter country codes (https://en.wikipedia.org/wiki/ISO_3166-1_alpha-3). If the study involves multiple countries, these are separated by “;”
Possible values:
- ISO 3 country code(s) - e.g.
DEU; DNKfor a study carried out in Germany and Denmark.
2.1.37 Continent (Population form)
Definition: Continent where study took place
Possible values:
AfricaAsiaEuropeN America- Includes Canada, USA & MexicoS America- Countries in the Americas except Canada, USA and MexicoOceania- Various definitions for Oceania exist, but here we opted for this one: http://en.wikipedia.org/wiki/List_of_Oceanian_countries_by_population
2.1.38 Ecoregion (Population form)
Definition: Indication of the ecoregion for the study, using the categories described in Figure 1 of Olson et al. (2001). For a more inclusive description of water ecoregions, see http://worldwildlife.org/biomes The exceptions for this are the codes LAB for studies conducted in laboratory or greenhouse conditions or NA for populations in which an assigning an ecoregion is not practical (e.g. human data pooled for an entire country)
Possible values:
TMB- Terrestrial - tropical and subtropical moist broadleaf forestsTDB- Terrestrial - tropical and subtropical dry broadleaf forestsTSC- Terrestrial - tropical and subtropical coniferous forestsTBM- Terrestrial - temperate broadleaf and mixed forestsTCF- Terrestrial - temperate coniferous forestsBOR- Terrestrial - boreal forests/ taigaTGV- Terrestrial - tropical and subtropical grasslands, savannas and shrublandsTGS- Terrestrial - temperate grasslands, savannas, and shrublandsFGS- Terrestrial - flooded grasslands and savannasMON- Terrestrial - montane grasslands and shrublandsTUN- Terrestrial – tundraMED- Terrestrial - Mediterranean forests, woodlands and scrubsDES- Terrestrial - deserts and xeric shrublandsMAN- Terrestrial – mangrovesLRE- Freshwater - large river ecosystemsLRH- Freshwater - large river headwater ecosystemsLRD- Freshwater - large river delta ecosystemsSRE- Freshwater - small river ecosystemsSLE- Freshwater - small lake ecosystemsLLE- Freshwater - large lake ecosystemsXBE- Freshwater - xeric basin ecosystemsPOE- Marine - polar ecosystemsTSS- Marine - temperate shelf and seas ecosystemsTEU- Marine - temperate upwellingsTRU- Marine - tropical upwellingsTRC- Marine - tropical coralLAB- Laboratory or greenhouse conditions – controlled, usually indoor, conditions that mean the study species is not affected by the environment conditions typical of the actual geographic location of the studyNA- Populations in which an assigning an ecoregion is not practical (e.g. human data pooled for an entire country)
2.1.39 StudiedSex (Matrix form)
Definition:
Possible values:
M- Studied only malesF- Studied only femalesH- Studied hermaphroditesM/F- Males and females separately in the same population matrix modelA- All sexes modeled together
2.1.40 MatrixComposite (Formerly Composition; Matrix form)
Definition: Indicates the type of matrix population model (also see Figure 2 below for more information)
Possible values:
Individual- A matrix population model constructed for a single study × species × population × treatment × period combinationMean- An average (mean) of other matrix population models (e.g. element-by-element arithmetic mean of a population’s matrices across several time periods available)Pooled- A matrix population model that has been constructed by pooling individual-level demographic information across populations and/or periods. This type of matrix, when available, has always been provided by the author either in the publication or through personal communications
2.1.41 MatrixSeasonal (Matrix form)
Definition: Indicates whether a matrix population model is seasonal, e.g., a matrix population model that does not describe a full annual transition, but rather a seasonal (< 1 yr) transition
Possible values:
Yes- The matrix population model is seasonalNo- The matrix population model is not seasonal
2.1.42 MatrixTreatment (Formerly Treatment; Matrix form)
Definition: Describes if a treatment was applied or not and the nature of the treatment
Possible values:
- treatment - A brief description of the treatment applied to the population described by the matrix population model. We define treatment as an action intentionally imposed by humans. If more than one applies to a matrix, treatments are separated with a “
;” Unmanipulated- No experimental, human-imposed treatment was applied. Natural events of non-intentional occurrence (e.g. fire, hurricanes) are recorded as “Unmanipulated”, but these incidences are described in the “Observations” variable (below)
2.1.43 MatrixCaptivity (Formerly Captivity; Matrix form)
Definition: Whether the study species was studied in the wild or under controlled conditions for most of its life cycle
Possible values:
W- Wild: study in natural conditionsC- Captive: studied for most part of the life cycle of the species in a botanical garden, green house, laboratory, etc.CW- Captured from Wild: If the species was taken from a wild population but studied in labs or gardens etc.
2.1.44 MatrixStartYear (Matrix form)
Definition: First year of study. Year t in annual matrix population model that describes population dynamics from time t to t+1.
Possible values:
- yyyy - e.g.
1994
2.1.45 MatrixStartSeason (Now MatrixSeasonal PLUS 2 more fields: MatrixStartSeason and MatrixEndSeaon; Matrix form)
Definition: First season of study
Possible values:
SpringSummerAutumnWinterNA
2.1.46 MatrixStartMonth (Matrix form)
Definition: First month of study
Possible values:
- 1-12 - e.g.
1= January,6= June, etc.
2.1.47 MatrixEndYear (Matrix form)
Definition: Last year of study. Year t+1 in matrix population model that describes population dynamics from time t to t+1
Possible values:
- yyyy - e.g.
1999
2.1.48 MatrixEndSeason (Matrix form)
Definition: Last season of study. Season s+1 in periodic matrix population model that describes population dynamics from season s to s+1. Here season is used as described in manuscript by the authors (Summer in the Southern Hemisphere corresponds to Winter in the Northern Hemisphere)
Possible values:
SpringSummerAutumn/FallWinterNA
2.1.49 MatrixEndMonth (Matrix form)
Definition: Last month of study
Possible values:
- 1-12 - e.g.
1= January,6= June, etc.
### CensusType
Definition: Sampling census used by the authors
Possible values:
Prebreeding- Population was censused before a birth pulsePostbreeding- Population was censused after a birth pulseIntermediate- Population was censused between successive birth pulses (e.g. bears give birth while hidden in winter dens, which makes pre- or post-breeding censuses impracticalBirth-flow- Population for which reproduction occurs continually over an interval (e.g. humans)
2.1.50 MatrixSplit (Matrix form)
Definition: This indicates whether the MPM A matrix has been divided into its constituent U, F and C submatrices. See Figure 3 below for further details
Possible values:
Divisible- The matrix population model A has been been divided into the process-based submatrices U, F and CIndivisible- The matrix population model A has not been divided into the process-based sub-matrices U, F and C because insufficient information is available to classify the various demographic processes for each sub-matrix. In indivisible matrices, only A has values given - the other matrices are filled withNAvalues
2.1.51 MatrixFec (Matrix form)
Definition: This records whether fecundity was measured at all for this matrix model
The rationale for this is that a fecundity (in the F matrix) may be recorded as 0 either because it was not measured, or because reproduction was measured and estimated to be zero. As a general rule, if any values for fecundity (e.g. in the F matrix) are non-zero, then MatrixFec should be recorded as ‘Yes’. If all values are zero, then you we check the paper carefully to check whther this is because no fecundity was detected, or whether no attempt was made to measure fecundity
Possible values:
Yes- Reproduction was included in the MPM, and thus values 0 should appear in the submatrix F if sexual reproduction occurred in these stages/ages. Note that it is possible that reproduction was measured and modelled but none occurred. In that case, values of 0 are possible in the F matrixNo- Reproduction was not modelled by the matrix population model, either because it was not the goal of the study or due to logistical impossibility. In some F submatrices the elements where sexual reproduction would have appeared containNAvalues, and in others they contain 0 valuesNA- it is not known whether fecundity was measured for the matrix
2.1.52 Observations (Matrix form)
Definition: This variable provides information that is not provided elsewhere but may nonetheless be important (e.g. plant canopy, burning intervals, etc.)
Possible values:
- any text - Any text string giving useful additional information
2.1.53 MatrixDimension (Matrix form)
Definition: Dimension of the matrix population model A
Possible values:
- integer - e.g.
5
2.1.54 SurvivalIssue (This is not a field, it needs to be calculated from the survival matrix)
Definition: This is a calculated field used for checking matrix data. It is the maximum stage-specific survival value in the survival U submatrix. To calculate SurvivalIssue, we sum each column in the U matrix individually. The maximum value among those sums is the SurvivalIssue.
Note: This value is only calculated with a number if MatrixSplit = “Divided”, that is, if the matrix is able to be split into submatrices. If MatrixSplit = “Indivisible”, the matrix is not divisible, so there is no survival U submatrix. In the case of indivisible matrices, the SurvivalIssue field will be empty (NA) and “Not Calculated” (NC) in the SQL database
Possible values:
- numeric - e.g.
0.9,1.2,1.45 NA- Matrix population model is not divisible into submatrices
2.1.55 _Database
Description: An internal check used for database creation and displays the database name (COMPADRE or COMADRE)
2.1.56 _PopulationStatus
Description: An internal check used for database creation. All values should display as Released
2.1.57 _PublicationStatus
Description: An internal check used for database creation. All values should display as Released
2.2 Variables in matrixClass
The object matrixClass is a list of data.frames detailing the class names used in each MPM
2.2.2 MatrixClassOrganized (Class Organized; Matrix form)
Definition: We standardize all stages in a given population matrix model to one of three stages (prop, active, dorm) to facilitate analyses
Possible values:
prop- Propagule (seed). This applies to every stage defined by the author as seed bank or seed. Users are encouraged to carefully examine matrices with these stages and to implement the appropriate calculations to avoid a spurious additional year being added when no seedbank exists in the study species, as explained by Caswell (2001, p. 60)active- This includes stages that can neither be placed in the prop nor dorm (see below) stagesdorm- A stage that is vegetatively dormant after having germinated and becoming established
2.2.3 MatrixClassNumber (The number of classes in a matrix. This has to be calculated from MatrixClassOrganized)
Definition: A numerical representation of classes in the population matrix model
Possible values:
- integer - e.g.
1,2,3, … n, where n is the dimension of the MPM (seeMatrixDimensionbelow)
2.3 Variables in mat
The object mat is a list of lists giving the actual MPMs in the form of the A matrix, and (where possible), the U, F and C matrices
2.3.1 matA (Matrix)
Description: The matrix population model A which describes the population dynamics of a population under conditions described by MatrixComposite, MatrixTreatment, MatrixStartYear, MatrixEndYear, Population, and Observation in the metadata
2.3.2 matU (Survival matrix)
Description: The population sub-matrix model U (a sub-matrix of A), which describes the survival-dependent dynamics of a population under the conditions described above. A range of numeric values are possible, but are constrained to be between 0 and 1. These include only survival-dependent vital rates (no sexual or clonal reproduction). The values are only digitized for Divided matrices (indicated by variable MatrixSplit)
2.3.3 matF (Fecundity matrix)
Description: The population sub-matrix model F (a sub-matrix of A), which describes the sexual-reproduction dynamics of a population under the conditions described above. A range of numeric values are possible, but are constrained to be positive. These include only estimates of sexual reproduction (no clonal reproduction).
The values are only digitized for Divided matrices (indicated by variable MatrixSplit)
2.3.4 matC (Clonal matrix)
Description: The population sub-matrix model C (a sub-matrix of A), which describes the clonal-reproduction dynamics of a population under the conditions described above. A range of numeric values are possible, but these are constrained to be positive. These include only estimates of clonal reproduction.
The values are only digitized for Divided matrices (indicated by variable MatrixSplit)
2.4 Variables in version
version is a list that provides some summary information.
2.4.1 Version
Description: The version number of the database (Major_Year_Month_Patch). Past versions used (Major_Minor_Patch) format. Numbering follows semantic versioning (www.semver.org)
2.4.2 Type
Description: The type of database release
Possible values:
Release- The standard COMPADRE or COMADRE releasePatch- A release version that has been patched since the previous standard release. Patches are released if there are significant additions or error corrections to the database between standard releasesTest- A version used strictly for internal testing of the databases. These versions do not follow semantic versioning, are not preserved indefinitely, and are not intended for public release
2.4.3 DateCreated
Description: Date of creation of the Rdata database object in the format MMM_DD_YYYY. E.g. Aug_06_2015
2.4.4 TimeCreated
Description: Time of creation of the Rdata database object in the format HH:MM. E.g. 19:08. Times are in GMT
2.4.5 Agreement
Description: The URL link for the data user agreement containing the open-access conditions to the use of these data (https://www.compadre-db.org/UserAgreement)
2.4.6 How to Cite
Description: The URL link for instructions on how to cite the data (https://compadre-db.org/Help/HowToCite)
2.4.7 GeneratorScriptVersion
Description: Version number for the code used to generate the COMPADRE and COMADRE databases. This version number is for internal tracking and troubleshooting
2.5 Appendix - The calculation of mean matrix population models
- In COMPADRE we present the individual matrix population models for each season, year, study population and treatment that can be calculated from a paper, as well as those personally communicated by the publication authors to the COMPADRE digitization team. When pertinent, mean matrix population models that are element-by-element averages (arithmetic mean) across all Unmanipulated matrix population models (defined above) and across each treatment group are also given. Mean matrix population models that combine estimates from different treatments, or that combine estimates from treatments with unmanipulated conditions, are not calculated because of their lack of biological interpretation (Figure 2).
- Seasonal matrix population models, where more than one matrix population model exists for a single year, are marked as “Seasonal”. For these, we calculate average matrix population models for each season but not for each year (Figure 3). Deriving an annual model from seasonal data is straightforward and the calculations are described by Caswell (2001, p. 349).
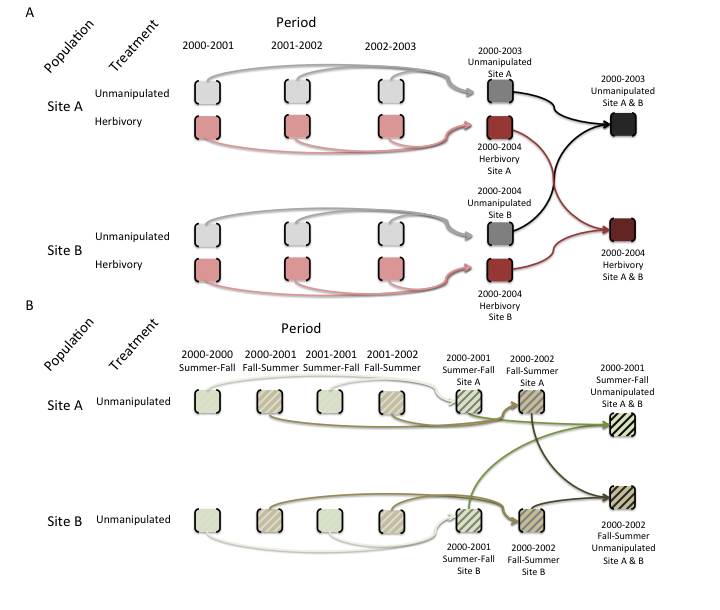
Figure 2. Schematic showing how mean population matrix models are derived from all available population matrix models in a study. A. For annual population matrix models (those that follow population dynamics from one year to the next, or sometimes more – some tree species are studied with a 5-year interval), element-by-element means are first calculated across the population matrix models describing the population dynamics for time periods within each population. Then the grand element-by-element mean is calculated across the matrix population models for the different populations. This procedure is repeated for models describing dynamics under each imposed treatment (pink matrices below) and for those describing the dynamics of unmanipulated populations (gray). Note that mean population matrix models are only calculated from population matrix models that share the same treatment (or absence of treatment). B. For seasonal or periodic population matrix models (e.g. for annual species), the mean seasonal population matrix model is calculated within populations and then across populations, both for matrices describing treated populations and for those describing populations with no imposed treatment (i.e. unmanipulated).
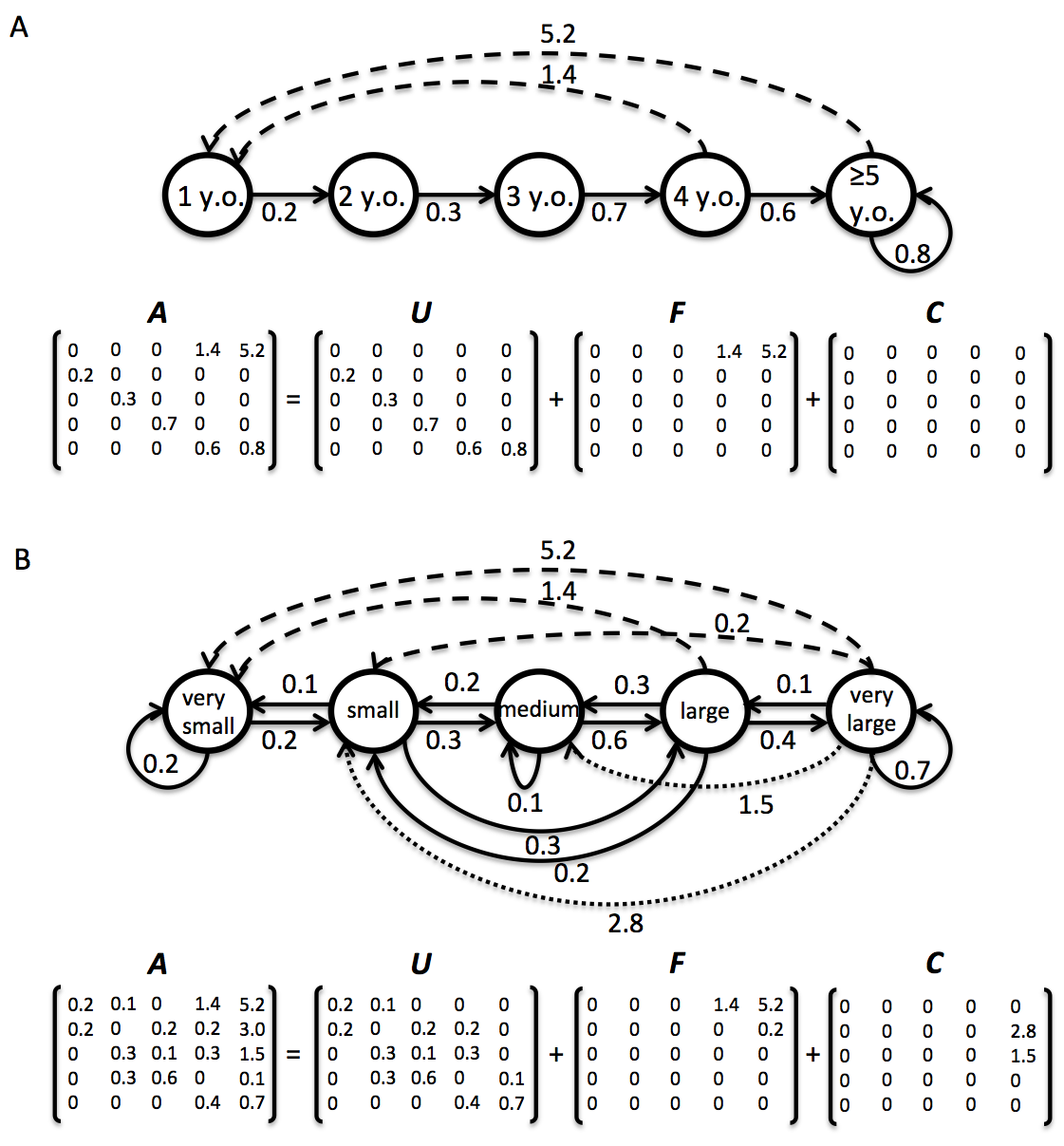
Figure 3. Life cycle of two hypothetical plant populations based on age (A) and size (B), with their corresponding matrix population models A, and underlying basic demographic processes of survival (U sub-matrix; solid arrows), sexual reproduction (F sub-matrix; dashed arrows) and clonal reproduction (C sub-matrix; dotted arrows). In the Leslie matrix model example (A) , the division of sub-matrices is more straightforward than in the Lefkovitch matrix model example (B). In the latter imaginary example, individuals can transition into the same stage as they can contribute with sexual and/or clonal offspring (e.g. small stage). In these cases, splitting A into sub-matrices U, F and C is only feasible when sufficient information is provided by the authors (see variable MatrixSplit in above).