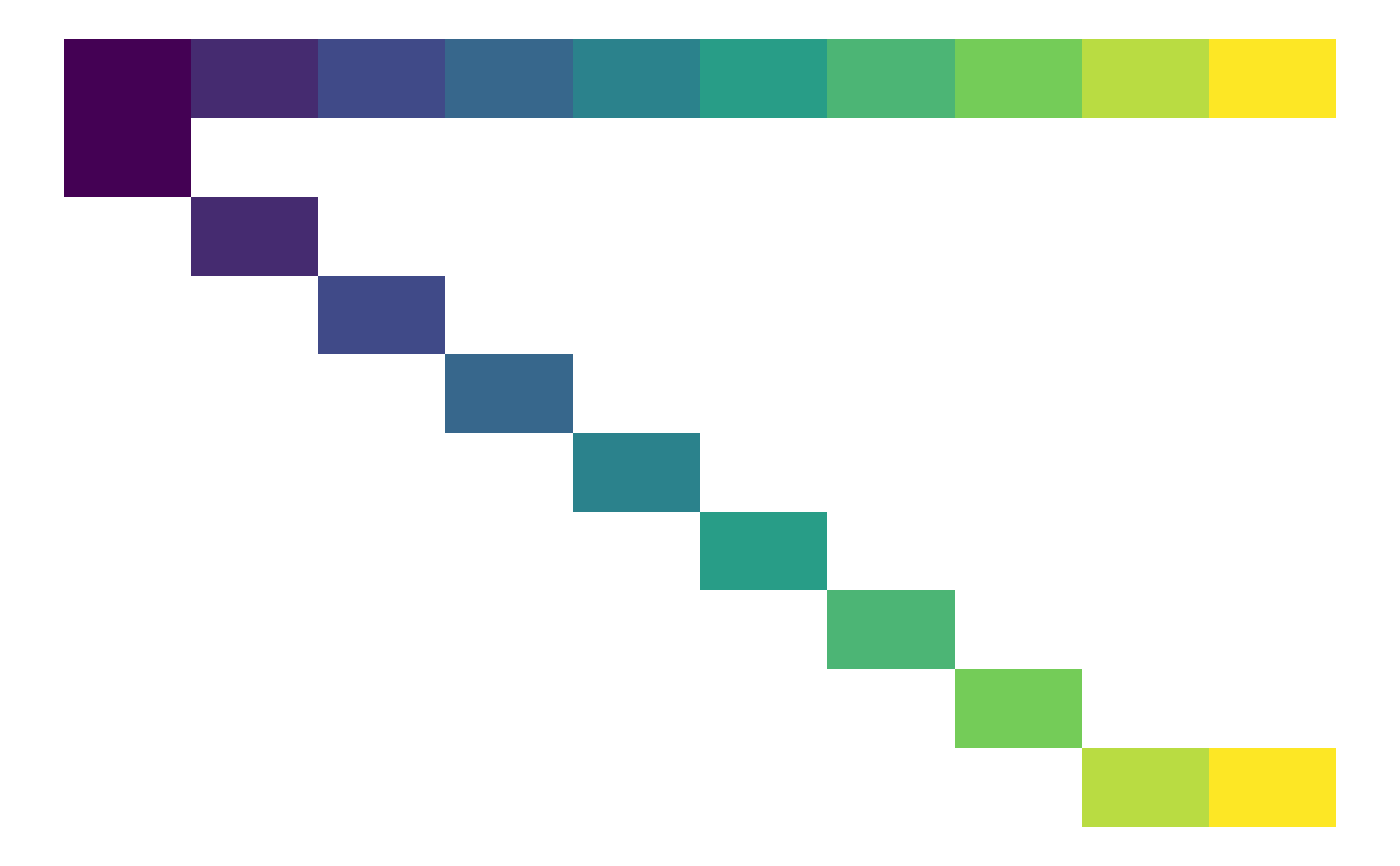Visualise a matrix, such as a matrix population model (MPM), as a heatmap.
See also
Other utility:
summarise_mpms()
Author
Owen Jones jones@biology.sdu.dk
Examples
matDim <- 10
A1 <- make_leslie_mpm(
survival = seq(0.1, 0.7, length.out = matDim),
fecundity = seq(0.1, 0.7, length.out = matDim),
n_stages = matDim
)
plot_matrix(A1, zero_na = TRUE, na_colour = "black")
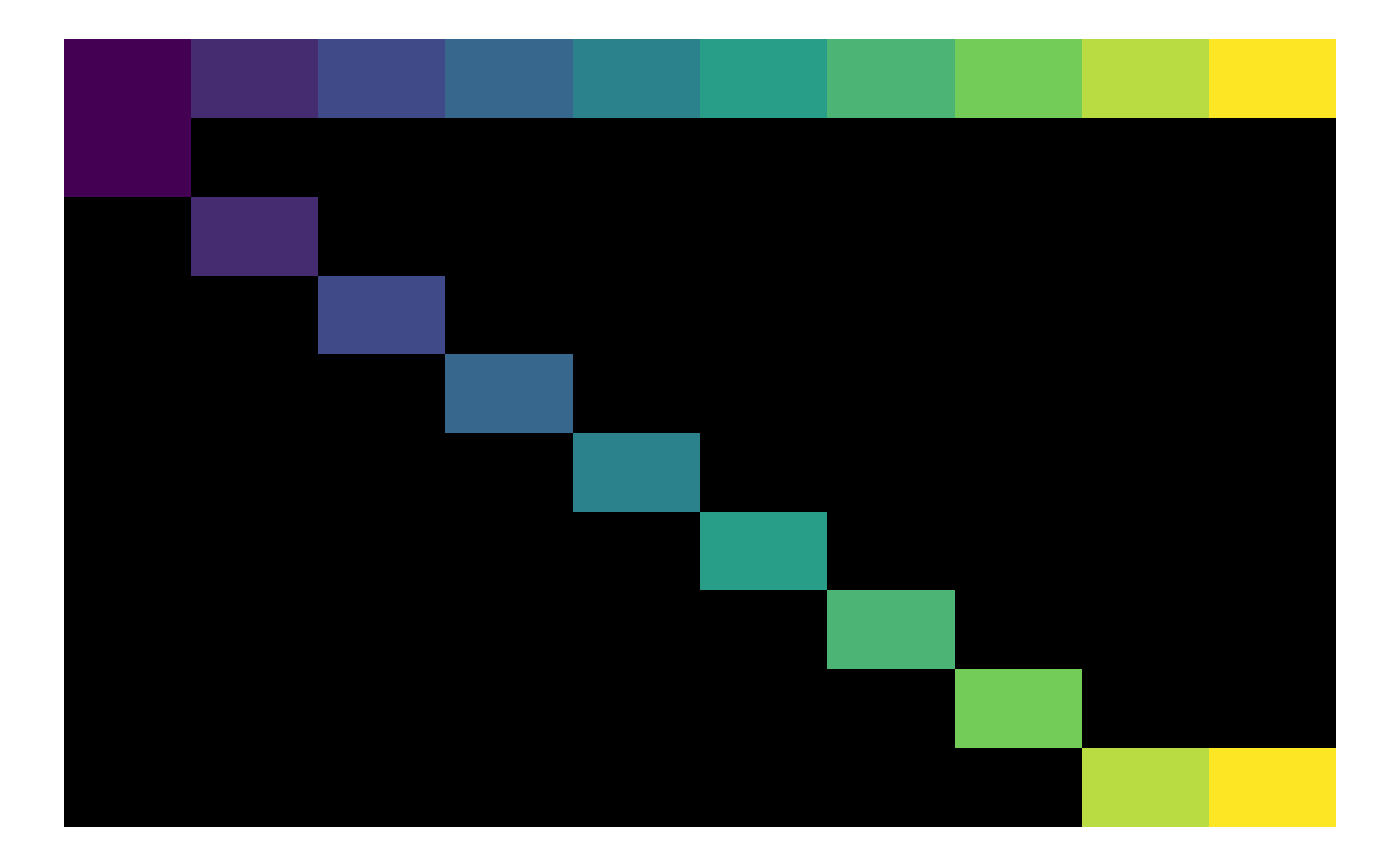 plot_matrix(A1, zero_na = TRUE, na_colour = NA)
plot_matrix(A1, zero_na = TRUE, na_colour = NA)